At Monstarlab we help build dedicated Machine Learning (ML) applications to tackle a client challenge that is best solved with this kind of technology. Within the ML domain, Deep Learning (DL) has been successfully applied to tackle data science challenges in many different areas. However, currently, the applications of DL in Life Sciences have been more limited.
In this article, we will describe how to use previously gathered, unrelated but large datasets to train a DL model that is then used to make predictions with a newer, smaller and dedicated dataset. This methodology is generally called Transfer Learning (TL). We also show how this can be embedded in a web app built with Streamlit in order to explore how the model performs on various tasks and datasets.
What's this about
It is difficult to find large, good-quality data to use in DL models. Depending on the number of parameters to train, which can be several millions, in order to train a DL model one often needs to have high-quality data consisting of thousands of samples. A rule of thumb for a pre-hoc determination of the number of samples needed to obtain a 95% accuracy from a DL model is 1000 samples for each category to predict.
Over the years, the scientific community in both private and public sectors have collected, curated and in some cases made publicly available datasets that the Machine Learning community routinely utilises to build and test models. What about using these data to tackle related challenges for which good data are hard to find?
In biotech and pharmaceutical industries, challenges for which data are hard to find include the prediction of molecule-to-molecule interactions, the cellular response to external stimuli such as drugs and antibiotics, or the prediction of cell growth and behavior. Here we demonstrate the use of public experimental gene expression data on cancer cell response to drugs applied to predict drug response of single cancer cells, for which adequate datasets are hard to find.
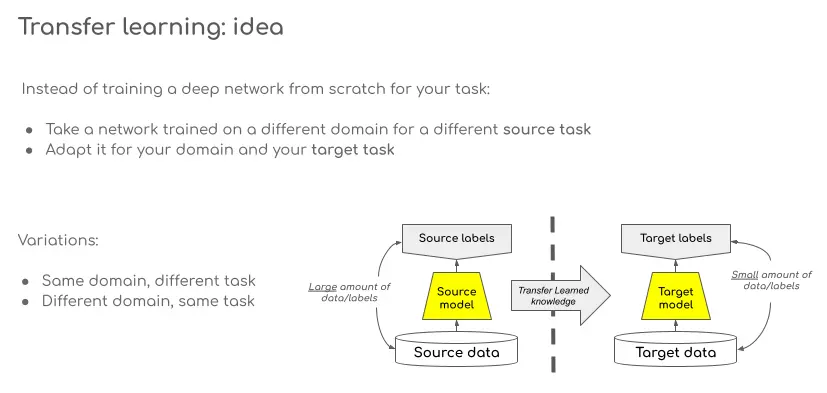
Transfer Learning: what it is and how it works.
Transfer learning (TL) is a research problem in machine learning (ML) that focuses on storing knowledge gained while solving one problem and applying it to a different but related problem. For example, knowledge gained while learning to recognize cars could be applied to recognize trucks. (Wikipedia)
Some advantages of using TL include a shorter training time and improved performance of DL models, as well as a reduced need of new and large training data. The way TL is adopted depends on the specific task at hand and factors such as whether the source and target data domains or tasks are the same or not.
TL is used to transfer knowledge learned with one dataset for a certain task to a model using a different dataset and/or tackling a different task. There are three basic ways to transfer this knowledge:
-
Inductive TL. In this type of TL, the source and target tasks are the same, however, are still different from one another. The model will use inductive biases from the source task to improve the performance of the target task. The source task may or may not contain labeled data, which leads to the model using multitask learning or self-taught learning respectively.
-
Unsupervised TL. In this case, the source and target data are similar and unlabeled, however, the tasks are different. Here the model trained on source data performs hopefully better in identifying patterns in target data. Dimensionality reduction and clustering are two of many unsupervised learning techniques. This approach can also be used for feature reduction if the target dataset is very large.
-
Transductive TL. In this type of TL, the source and target tasks share similarities, however, the domains are different. For example, the source domain contains a lot of labeled data while the target domain does not. Here TL is used for domain adaptation.
Cancer cell lines and single cells
The use of single-cell omics data has seen in recent years a very strong interest in the pharmaceutical industry. The availability of new technology and computational and bioinformatics methods allow now to obtain a much larger amount of data at a much higher resolution.
Single-cell RNA-sequencing (scRNA-seq) data are very valuable to dissect the individual response of cancer subpopulations to specific drugs. However, public, good quality and benchmarked single-cell data that can be used to train DL models are limited. On the other hand, abundant in vitro drug screening studies have been conducted over the years, leading to the availability of drug response data on different cancer cell lines (CCLs).
CCls bulk RNAseq data can be a great resource to help train DL models that predict drug responses at single-cell level based on scRNA-seq data. Here we show how this can be done via TL.
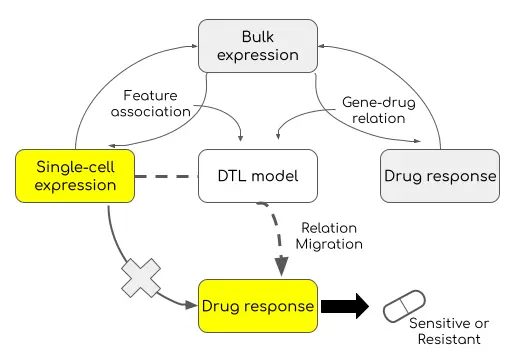
Transfer Learning applied to RNAseq data
We present here a framework published recently by a team of academic researchers under the name scDEAL (J. Chen et al., 2021). The framework involves the following 4 major components:
- a bulk gene feature extractor
- a single-cell gene feature extractor
- a drug response predictor
- a whole Deep Transfer Learning (DTL) model combining all the extractors and the predictor as one.
Without going too much into the details, the extractors extract gene expression features that best represent the original datasets. The predictor is trained to receive bulk RNAseq data and minimize the difference between predicted and true drug response labels. The whole pre-training of these models also aims at generating the best Neural Network weights to be used within the DTL model. This model works in a multitask (Inductive TL) manner by minimizing the differences in gene features between the two extractors, as well as the loss function of the common predictor output.
What is Streamlit
Streamlit is an open-source Python library that makes it easy to create custom web apps for ML and data science. Streamlit is an elegant and simple Python-only library, which helps reduce the time needed to make a web app from weeks to hours especially when the ML framework is also developed in Python. Streamlit has seen a rapid expansion since its inception and has garnered a large and bustling community of developers and enthusiasts.
Based on the scDEAL framework and the available RNAseq data, we were able to quickly build a Streamlit app enabling us to run and test different versions of the DTL model, using different parameters and different encoder/decoder pairs.
Through the web app, changing and running the model, and explore the results, was done by simply using buttons and dropdown menus. In other words, a web app allows a scientist with no coding experience to explore and test ML and DL applications very easily.
Explore DTL model predictions
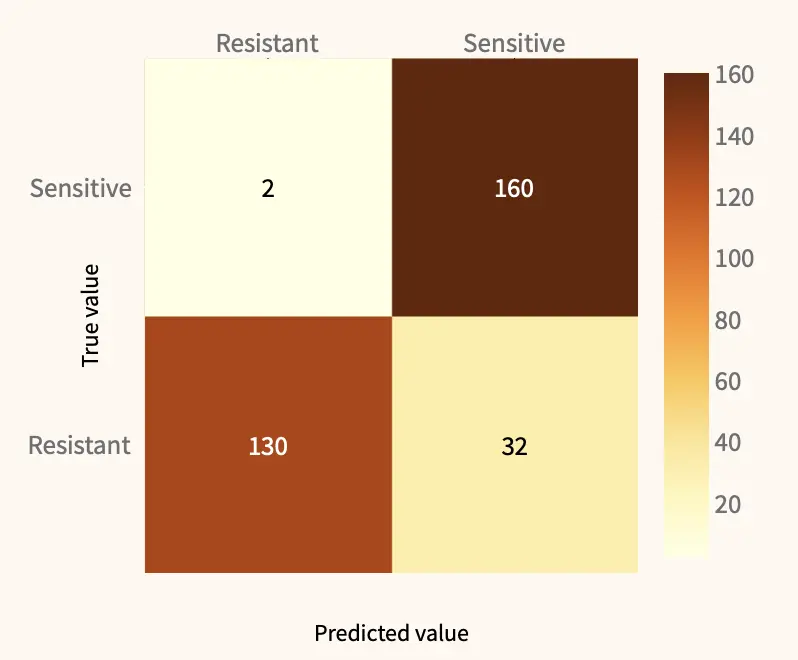
We tested one DTL model in its ability to predict resistant and sensitive single cells based on scRNAseq data. The scDEAL framework covers many different types of CCL and drug combinations. All these combinations were integrated into our Streamlit app, allowing for an easy exploration of all possible combinations, and calculating performance metrics of the DTL model.
Precision and recall, two very popular performance metrics, can be calculated based on either the number of correctly called drug-sensitive or resistant cells. For example, when predicting the response of prostate cancer cells to the drug Docetaxel, the model showed high prediction performance of drug-sensitive cells with a 0.83 precision and 0.98 recall.
Conclusion
The main obstacle in developing DL tools for predicting single-cell drug responses, and for other tasks in biological sciences, is the lack of sufficient training power due to the limited number of benchmarked data in the public domain.
Here, we demonstrate how DTL can be used to integrate and trasfer knowledge from cell line to single-cell data. Additionally, we show how this modeling framework can be embedded in an easy-to-use web application for researchers with little or no DL or coding knowledge.



